Hi Eric,
I am back from my trip. Here is what your data look like using a Delaunay triangulation to rebuild the underlying mesh:
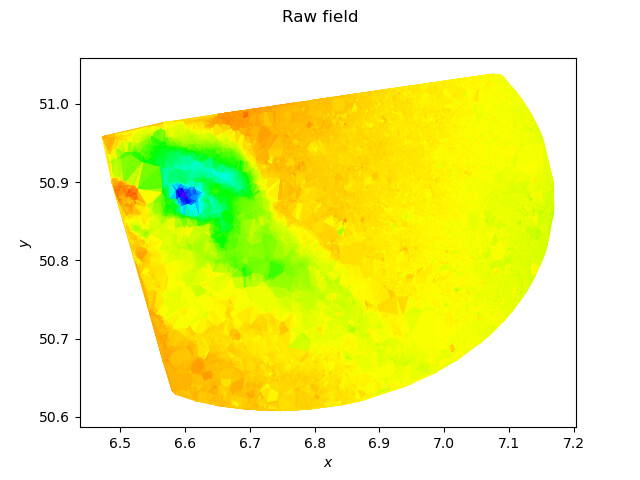
Using a P1 interpolation:
import openturns as ot
import openturns.viewer as otv
import numpy as np
from scipy.spatial import Delaunay
sample = ot.Sample.ImportFromTextFile('cleandata.csv', ',', 2)
points = sample.getMarginal([1, 2])
visuMesh = False
print("Build triangulation")
tri = Delaunay(points)
print("Build mesh")
mesh = ot.Mesh(points, [[int(x) for x in t] for t in tri.simplices])
if visuMesh:
import matplotlib.pyplot as plt
plt.triplot(points[:,0].asPoint(), points[:,1].asPoint(), tri.simplices)
plt.plot(points[:,0], points[:,1], 'o')
plt.show()
ot.ResourceMap.SetAsUnsignedInteger("Mesh-LargeSize", 0)
ot.Show(mesh.draw())
field = ot.Field(mesh, sample.getMarginal(3))
print("Display raw field")
g = field.draw()
g.setXTitle(r"$x$")
g.setYTitle(r"$y$")
g.setTitle("Raw field")
view = otv.View(g)
view.save("field.png")
view.close()
print("Min/Max values")
print(field.getValues().getMin(), field.getValues().getMax())
print("Build P1 interpolation")
function = ot.Function(ot.P1LagrangeEvaluation(field))
xyMin = points.getMin()
xyMax = points.getMax()
M = 256
square = ot.IntervalMesher([M]*2).build(ot.Interval(xyMin, xyMax))
fieldP1 = ot.Field(square, function(square.getVertices()))
g = fieldP1.draw()
g.setXTitle(r"$x$")
g.setYTitle(r"$y$")
g.setTitle("Interpolated field")
view = otv.View(g)
view.save("fieldP1.png")
view.close()
you get:
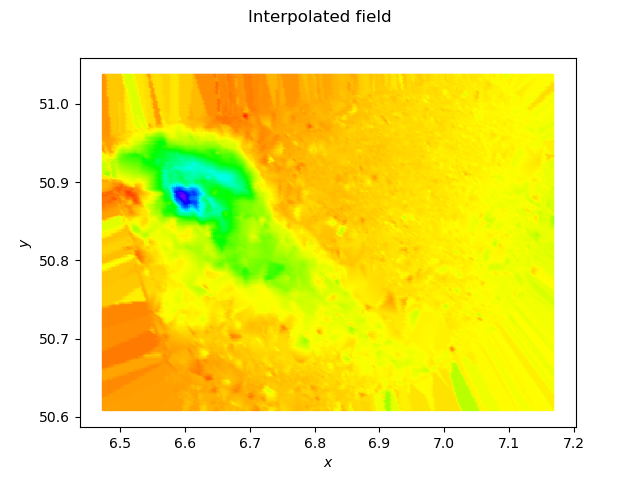
You see the blurry effect due to the linear interpolation, and the rays outside of the mesh due to the extension of the piecewise linear function by a piecewise constant function in these regions.
I also tried to use KrigingAlgorithm with HMAT on your data. Unfortunately there is a bug with SphericalModel and I get an exception. Switching to a MaternModel with nu=3/2 to get approximately the same level of smoothness I get the following computation time, depending of the size of the subsample I use:
# Matern 3/2 oss
# 1000 0.9618020057678223
# 10000 20.189648628234863
# 20000 48.61438536643982
# 50000 399.1733877658844
# 100000 1231.114249944687
# full 1172.22917294502264
so you see that it works (i.e. it fits into the memory and it returns an answer in a finite amount of time) but it is expensive, and the resulting meta-model is slow. The output is:
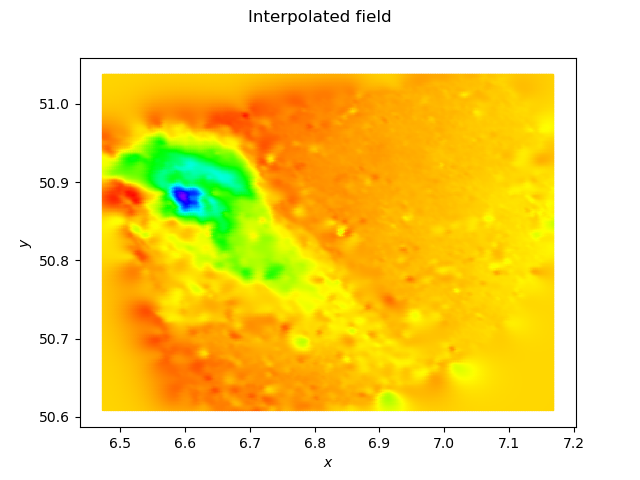
which is quite similar to the P1 result on the initial mesh, and a little bit smoother outside of the mesh. The script I used is:
import openturns as ot
import openturns.viewer as otv
import numpy as np
import time
# Here we use numpy to transpose data
sample = ot.Sample.ImportFromTextFile('cleandata.csv', ',', 2)
points = sample.getMarginal([1, 2])
dim = points.getDimension()
z = sample.getMarginal(3)
delta = points.computeRange()
xyMin = points.getMin()
xyMax = points.getMax()
print("points=", points.getSize())
print("z=", z.getSize())
print("delta=", delta)
# Spherical kernel
#kernel = ot.SphericalModel(2)
#kernel.setActiveParameter([0,1,2,3])
#kernel.setParameter([0.1]*4)
kernel = ot.MaternModel([0.1]*2, [1.0], 1.5)
#kernel.setActiveParameter([])
#kernel.setParameter([0.1, 0.1, 1.0, 1.5])
ot.ResourceMap.SetAsString("KrigingAlgorithm-LinearAlgebra", "HMAT")
ot.ResourceMap.SetAsScalar("HMatrix-AssemblyEpsilon", 1e-6)
ot.ResourceMap.SetAsScalar( "HMatrix-RecompressionEpsilon", 1e-7)
algo = ot.KrigingAlgorithm(points, z, kernel, ot.Basis())
optimizer = ot.NLopt("LN_COBYLA")
algo.setOptimizationAlgorithm(optimizer)
ot.Log.Show(ot.Log.INFO)
t0 = time.time()
algo.run()
t1 = time.time()
print("t=", t1 - t0, "s")
metaModel = algo.getResult().getMetaModel()
M = 256
square = ot.IntervalMesher([M]*2).build(ot.Interval(xyMin, xyMax))
fieldP1 = ot.Field(square, metaModel(square.getVertices()))
g = fieldP1.draw()
g.setXTitle(r"$x$")
g.setYTitle(r"$y$")
g.setTitle("Interpolated field")
view = otv.View(g)
view.save("fieldKriging.png")
view.close()
Cheers
Régis







 and have a great trip! I did hike in the alps a couple of years ago too.
and have a great trip! I did hike in the alps a couple of years ago too.




